Combating new viral outbreaks in the 21st century
The explosive emergence of Zika in the past two years was a sobering reminder that relatively obscure viruses can arise suddenly as global health emergencies. In the Flaviviridae alone, the family of viruses that Zika belongs to, there are more than 20 viruses with the potential to cause disease in humans. Unfortunately, there is no yet a satisfactory way to predict and stop the next epidemic, so we will likely remain susceptible to new emergent infections. The best-known antidote against viral spread is vaccination, but the road to the designing, testing, and implementing effective vaccines is notoriously protracted. In fact, it could be decades before we have a Zika vaccine licensed for use; a rather delayed response to an outbreak that started in 2014. How can we quickly respond to new viral challenges then? Fortunately, it may be possible to shorten the future development of preventative therapies with promising new technologies to block infections. Our research group–in fact, an expanding community of researchers–has devoted a lot of effort to find molecules that could bridge this development gap, and the field is now enthused with the potential of special proteins called ‘monoclonal antibodies’.
The body usually responds to infections by creating several diverse antibodies that can bind specifically to the invading microbes and stop pathogen replication. Inducing these protective antibody responses has been the goal of vaccine research for almost every microbial disease. The recent, not so quiet revolution in the field, is that we can now isolate individual clones of cells producing antibodies. Each clone has the recipe to produce a single antibody, and copying these recipes–hence the name ‘monoclonal’– allows us to recreate the molecules that compose the overall response to a given pathogen. In more practical terms, it is now possible, for example, to obtain blood from a few individuals that had Zika infections and isolate monoclonal antibodies that can block the Zika virus. The hope is that, in an event of a new outbreak, these molecules can be ready to be administered early to populations at risk and limit the spread.
We have tested the concept of using monoclonal antibodies instead of traditional vaccines; the results were recently published in the journal Science Translational Medicine. We engineered three monoclonal antibodies against Zika virus based on sequences from a Zika patient. Administration of a cocktail with these antibodies completely prevented macaques from becoming infected with Zika virus. Thus, our results strongly suggest that passive immunization with a monoclonal antibody cocktail with might be also effective in humans.
For the foreseeable future, the prevention and treatment of emerging infectious diseases will continue to be of major international significance. Monoclonal antibodies are a recent and promising tool for our playbook against microbial diseases. These are exciting times, while we are vulnerable to emerging infectious pathogens, the combination of technologies for early pathogen detection, and early development of experimental therapies might lead to complete novel manners to stop epidemics–Hopefully, before they even become epidemics.
https://www.nature.com/nature/journal/v546/n7658/full/nature22400.html
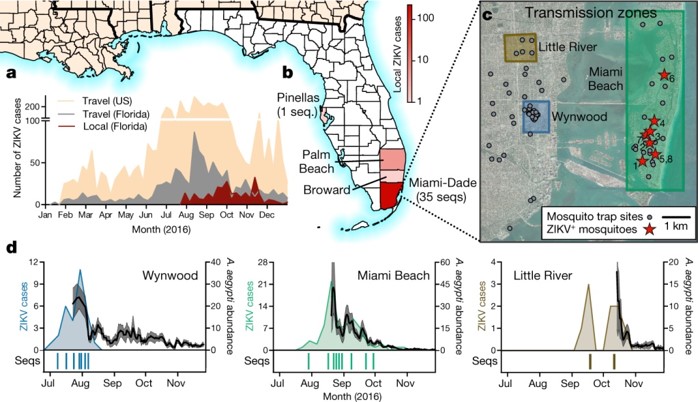
a, Weekly counts of confirmed travel-associated and locally acquired ZIKV cases in 2016. b, Four counties reported locally acquired ZIKV cases in 2016: Miami-Dade (241), Broward (5), Palm Beach (8), and Pinellas (1). There was also one case of unknown origin. c, The locations of mosquito traps and collected A. aegypti mosquitoes found to contain ZIKV RNA (ZIKV+) in relation to the transmission zones within Miami. d, Temporal distribution of weekly ZIKV cases (left y-axis), sequenced cases (bottom), and A. aegypti abundance per trap night (right y-axis) associated with the three described transmission zones. ZIKV cases and sequences are plotted in relation to symptom onset dates (n = 18). Sequenced cases without onset dates or that occurred outside the transmission zones are not shown (n = 10). Human cases and A. aegypti abundance per week were positively correlated (Spearman r = 0.61, Extended Data Fig. 1b). The maps were generated using open source basemaps (http://www.esri.com/data/basemaps).